1. Does the direction in which a sequence is replicated affect the consensus of its USSs? That is, do USSs whose "forward" orientation sequence is found in the leading strand during DNA replication have a slightly different consensus than those whose "reverse" orientation sequence is in the leading strand? I raised this issue at the end of an earlier post, but I haven't tested it yet. All I need to do is to get the sequence of each strand of the genome, chop them at the origin and terminus of replication, and then put the parts together so all the leading strand sequences are in one file, and all the lagging strand sequences are in another file. Then run both files through the motif search.Yesterday I finally set out to test this.
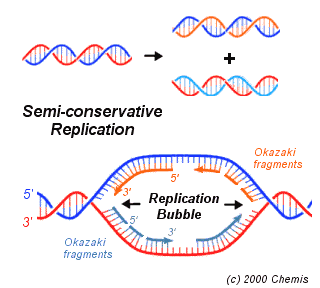 I had expected that the tricky step would be assembling the parts of the genome sequence into the two files, and I was right. We teach the concept of DNA having two antiparallel strands as if it was simple ("5' end, 3' end, how hard can this be?") but it's full of traps for the unwary. I had made (on the hall whiteboard and in my notebook during a boring seminar) preliminary sketches of the relationships between the physical strands of the genome, the location of the origin and terminus of replication, the structure of the bidirectional replication forks, and the 'forward' and 'reverse complement' DNA sequences available from TIGR. But I still got muddled and had to supplement my fresh pencil sketches with a new complex multi-coloured drawing on the whiteboard and an attempt to explain it to a passing post-doc. (The image is taken from here.)
I had expected that the tricky step would be assembling the parts of the genome sequence into the two files, and I was right. We teach the concept of DNA having two antiparallel strands as if it was simple ("5' end, 3' end, how hard can this be?") but it's full of traps for the unwary. I had made (on the hall whiteboard and in my notebook during a boring seminar) preliminary sketches of the relationships between the physical strands of the genome, the location of the origin and terminus of replication, the structure of the bidirectional replication forks, and the 'forward' and 'reverse complement' DNA sequences available from TIGR. But I still got muddled and had to supplement my fresh pencil sketches with a new complex multi-coloured drawing on the whiteboard and an attempt to explain it to a passing post-doc. (The image is taken from here.)I double-checked the final files by making sure that the 8 or 10 bases at the end of each were complementary to what I thought should be the corresponding end of the other file's sequence, and they were, so I think I've got it right. But I should triple-check then today, especially if the answer looks interesting.
The sequence files then had to be massaged into a form the motif-search program could handle: get rid of carriage returns, remove unacceptable characters (AGCTN are OK, others aren't), chop into 9kb segments). Throw out files and repeat, this time removing the extra bits of identifying text first. Throw out files and repeat, typing commands correctly (curse you, command line interface!).
And then the motif-search program scorned my sequences! The problem appeared to be the same one I was having with my gene-sequence files (next post); the program would appear to be processing the sequences as it should, but at the end it would shrug and say "sorry, I didn't find any patterns", even though I knew the patterns were there. Suspicion centered on the carriage returns, so I threw out the files and did everything again.
Success! The files were acceptable, and the motif search appeared to be actually searching, rather than just going through the motions. So I set up two runs, one with the leading strand sequences and one with the lagging strand sequences, and later this morning I should have the results.
















No comments:
Post a Comment
Markup Key:
- <b>bold</b> = bold
- <i>italic</i> = italic
- <a href="http://www.fieldofscience.com/">FoS</a> = FoS